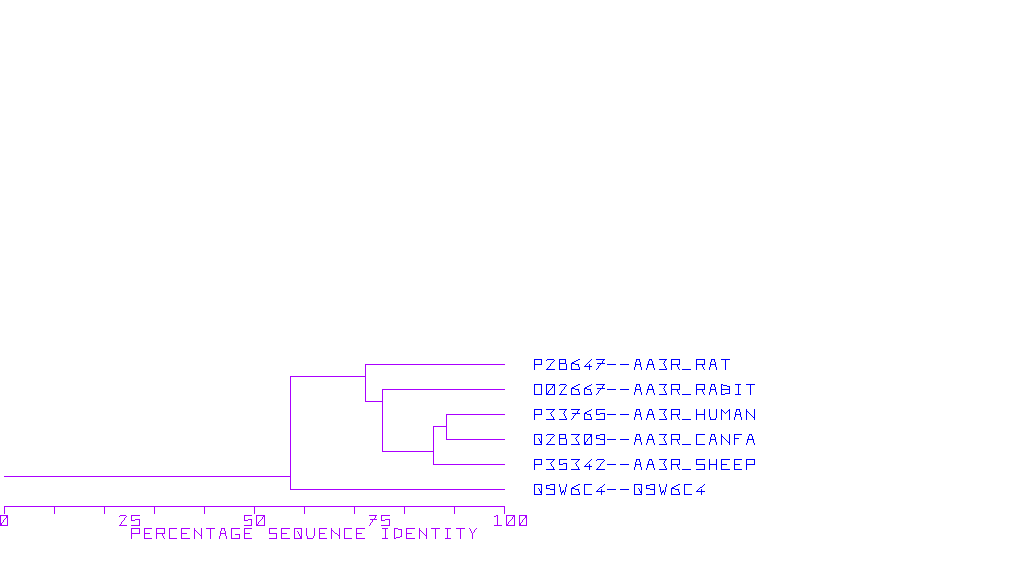
Adenosine Receptor A2a
![]()
The A2b adenosine receptor (A2aAR) has been cloned from a variety of species including canine, rat, and human (Olah and Stiles, 1995). A2aARs are typically larger than the A1ARs and range in size from 410- 412 amino acids corresponding to a protein of about 45, 000 daltons. The most striking difference between A1 and A2a is in the size of the carboxyl tail; the A2a tail being much longer than the carboxyl terminal tail found in A1ARs. The presence in the tail of multiple serines and threonines suggests that this region may be important in the regulation of the receptor by phosphory;lation following agonist stimulation. The A2b adenosine receptor was cloned from a human hippocampal cDNA library in 1992 (Furlong et al., 1992). This receptor is positively coupled to adenylate cyclase and displays high affinity binding for the A2a specific agonist CGS21680.
This, however, seems unlikely, since mutated canine A2a in which stop codons were inserted at amino acids 309 or 316 lead to markedly truncated carboxyl terminal tails as defined by photoaffinity labeling. These mutations failed to alter either the binding of ligands to this receptor or the receptor's ability to stimulate adenylyl cyclase. Further studies will be needed to understand the importance of this unique structural feature of A2a receptors.
Site-
directed mutagenesis studies
Databases of A2a Adenosine Receptor
| CFGPCR8 | A2A-Adenosine Receptor | Canis familiaris | |
| S105722 | A2A-Adenosine Receptor | Rattus norvegicus | |
| HUMA2XXX | A2A-Adenosine Receptor | Homo sapiens | |
| RATADENO | A2A-Adenosine Receptor | Rattus norvegicus | |
| S46950 | A2A-Adenosine Receptor | Homo sapiens | |
| HSA2AREC | A2A-Adenosine Receptor | Homo sapiens | |
| CPU04201 | A2A-Adenosine Receptor | Cavia porcellus | |
| MMU05672 | A2A-Adenosine Receptor | Mus musculus | |
| HSA2AAR01 | A2A-Adenosine Receptor | Homo sapiens | |
| GPIAA2AR | A2A-Adenosine Receptor | Cavia porcellus | |
| HSHA2AR01 | A2A-Adenosine Receptor | Homo sapiens | |
| MMADORA2A | + | A2A-Adenosine Receptor | Mus musculus |
| MMA2AEX1 | + | A2A-Adenosine Receptor | Mus musculus |
For up to 20 representative sequences for A2a and A2b adenosine receptors
![]()